Simulation Exercise
Exercise on simulating data using the simulate.py script and performing family-based polygenic score analysis.
Simulating data
If snipar has been installed succesfully, the command line scripts should be accessible as executables in your terminal. snipar includes a script for simulating genotype-phenotype data according to different scenarios: direct and indirect genetic effects, with and without assortative mating. To simulate data, please first create a directory to store the data:
mkdir sim
Now, we are going to simulate data for 3000 families, each with two full-siblings, genotyped at 1000 independent SNPs. We simulate a phenotype affected by direct genetic effects and assortative mating. We are going to simulate 20 generations of assortative mating with parental phenotype correlation 0.5, reaching an approximate equilibrium. The command for this is:
simulate.py 1000 0.5 sim/ --nfam 3000 --impute --n_am 20 --r_par 0.5 --save_par_gts
where the first argument gives the number of causal SNPs, the second argument gives the random mating heritability, the third gives the output directory, –nfam gives the number of families, –impute tells the script to impute missing parental genotypes, –n_am gives the number of generations of assortative mating, the parental phenotype correlation is given by –r_par, and –save_par_gts tells the script to output the genotypes of the parents of the final generation in addition to the genotypes of the final generation.
Please change your working directory to sim/:
cd sim
In this directory, the file phenotype.txt is a phenotype file containing the simulated phenotype.
The genotype data (chr_1.bed) has been simulated so that there are 3000 independent families, each with two siblings genotyped.
Inferring IBD between siblings
The first step is to infer the identity-by-descent (IBD) segments shared between siblings. However, for the purpose of this simulation exercise (where SNPs are independent, so IBD inference doesn’t work) we have provided the true IBD states in the file chr_1.segments.gz.
Imputing missing parental genotypes
This is performed using the impute.py script. To impute the missing parental genotypes without using phase information, use this command:
impute.py --ibd chr_@ --bed chr_@ --pedigree pedigree.txt --out chr_@ --threads 4
The pedigree along with the IBD segments shared between siblings recorded in chr_1.segments.gz are used to impute missing parental genotypes from the observed sibling and parental genotypes in chr_1.bed. The imputed parental genotypes are output to a HDF5 file, chr_1.hdf5.
Polygenic score analyses
snipar provides a script (pgs.py) for computing polygenic scores (PGS) based on observed/imputed genotypes, and for performing family based polygenic score analyses. The script computes a PGS from a weights file.
To compute the PGS using the true direct genetic effects as weights, use the following command:
pgs.py direct --bed chr_@ --imp chr_@ --weights causal_effects.txt --beta_col direct
It outputs the PGS to a PGS file: direct.pgs.txt. The pgs computation script automatically estimates the correlation between parents PGS values (also using full-sibling offspring PGS values to do this) and performs an adjustment for assortative mating when using the imputed parental genotypes to compute the PGS.
To estimate direct effect and average NTC of the PGS, use the following command:
pgs.py direct --pgs direct.pgs.txt --phenofile phenotype.txt
This will output a population effect estimate (1 generation model) to direct.1.effects.txt, and direct effect and average NTC estimates to (2 generation model) to direct.2.effects.txt. The population and direct effect estimates are the coefficients on the proband PGS in the 1 and 2 generation models, so are indicated by the ‘proband’ row. The average NTC estimate is the coefficient on the parental PGS in the two-generation model. The first column gives the name of the covariate/PGS column, the second column gives the estimated regression coefficient, and the third column gives the standard error of the estimate. The sampling variance-covariance matrix of the estimates is output to direct.1.vcov.txt (for the 1 generation model) and direct.2.vcov.txt (for the 2 generation model).
As we are using the true direct effects as weights, the PGS captures all of the heritability, and the direct and population effects should both be the same (1 in expectation), and the average parental NTC should be zero (in expectation). To check this, read in the effect estimate output files in R or look at them using a text viewer (e.g. less -S on a unix system).
To compute the PGS from the true direct genetic effects+estimation error (such as would be obtained from a family-based GWAS), use the following command:
pgs.py direct_v1 --bed chr_@ --imp chr_@ --weights causal_effects.txt --beta_col direct_v1
It outputs the PGS to a PGS file: direct_v1.pgs.txt. (Notice also that the inferred correlation between parents’ PGSs is lower than when using the true direct genetic effects as weights due to estimation error in the weights.)
To estimate direct effect and average NTC of the PGS, use the following command:
pgs.py direct_v1 --pgs direct_v1.pgs.txt --phenofile phenotype.txt
This will output a population effect estimate (1 generation model) to direct_v1.1.effects.txt, and direct effect and average NTC estimates to (2 generation model) to direct_v2.2.effects.txt.
Unlike when using the true direct genetic effects as weights, the direct effect of the PGS estimated from noisy weights (in direct_v1.1.effects.txt) will be smaller than the population effect (direct_v1.2.effects.txt). This is because the PGS does not capture all of the heritability due to estimation error in the weights. The PGS has its population effect inflated (relative to its direct effect) by assortative mating, which induces a correlation of the PGS with the component of the heritability not directly captured by the PGS due to estimation error. This inflation is not captured by the direct effect of the PGS because of the within-family variation used to estimate the direct effect is due to the random segregation of genetic material during meiosis. Here, the ratio between direct and population effects of the PGS should be around 0.86.
One should also observe a statistically significant average parental NTC (in direct_v1.2.effects.txt) of the PGS from the two-generation model despite the absence of parental indirect genetic effects in this simulation. Here, the ratio between the average NTC and the direct effect should be around 0.15. This demonstrates that statistically significant average NTC estimates cannot be interpreted as demonstrating parental indirect genetic effects, especially for phenotypes affected by assortative mating.
Adjusting for assortative mating
We now show how to adjust two-generation PGI results for assortative mating using the procedure outlined in Estimation of indirect genetic effects and heritability under assortative mating. The estimation procedure is summarized in this diagram:
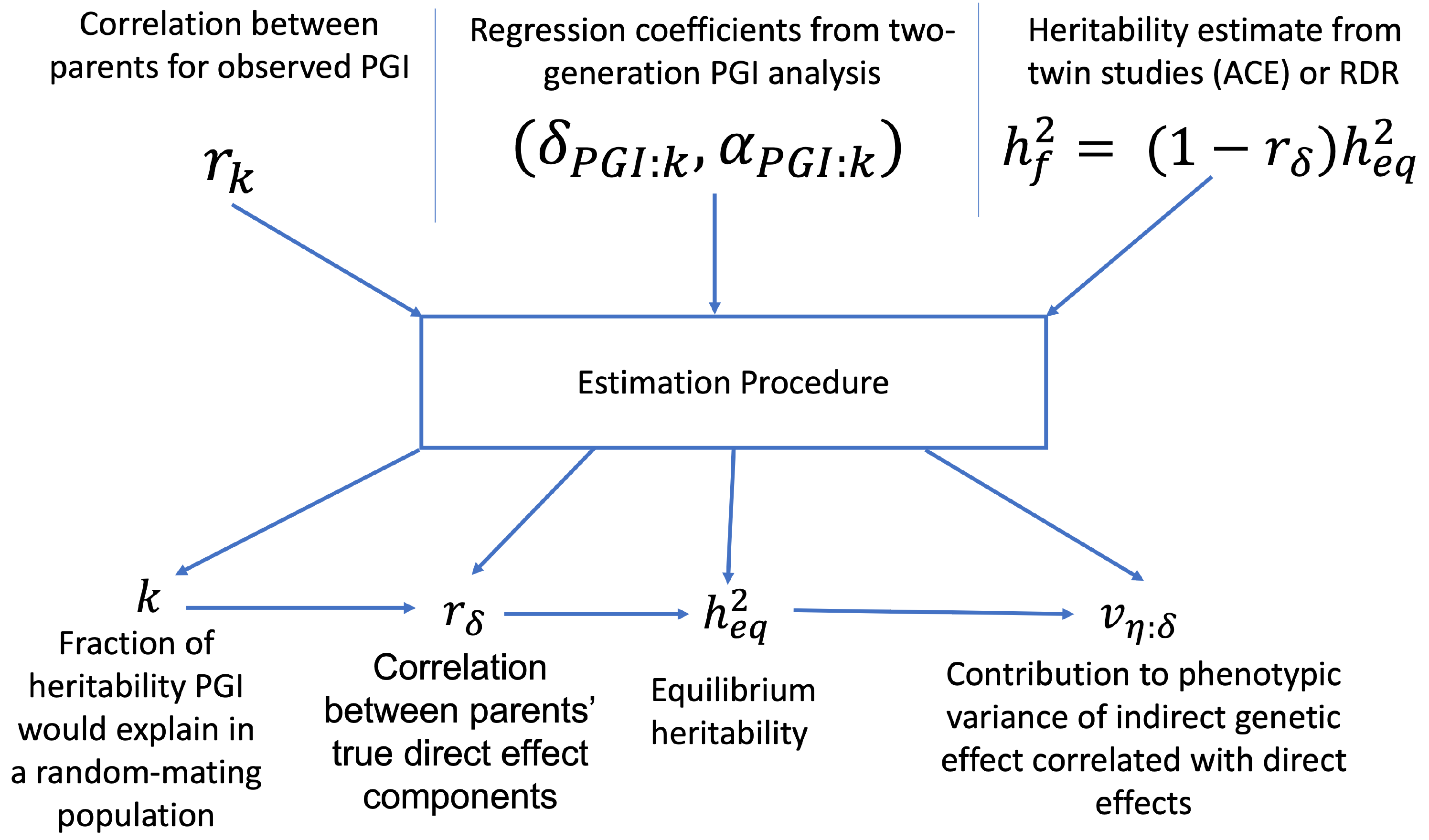
The estimation requires as inputs: an estimate of the correlation between parents’ scores, \(r_k\); the regression coefficients from two-generation PGI analysis, (\(\delta_{\text{PGI}:k},\alpha_{\text{PGI}:k}\)); and a heritability estimate, \(h^2_f\),from MZ-DZ twin comparisons, RDR, or sib-regression.
The estimation procedure outputs estimates of: \(k\), the fraction of heritability the PGI would explain in a random mating population; \(r_\delta\), the correlation between parents’ true direct genetic effect components; \(h^2_\text{eq}\), the equilibrium heritability, adjusting for the downward bias in heritability estimates from MZ-DZ comparisons, RDR, and sib-regression; \(\alpha_\delta\), the indirect genetic effect of true direct genetic effect PGI; and \(v_{\eta:\delta}\), the fraction of phenotypic variance contribued by the indirect genetic effect component that is correlated with the direct genetic effect component.
We can use snipar to compute the two-generation PGI estimates and the correlation between parents’ scores, and we can input a heritability estimate into pgs.py script to complete the inputs, so that snipar will perform the two-generation analysis adjusting for assortative mating.
To perform the estimation, we will combine the offspring and parental genotype files. This enables us to estimate the correlation between parents’ scores using the observed parental genotypes. (This is better than using the sibling genotypes because the correlation estimate from observed parental genotypes is uncorrelated with the PGS regression coefficients.)
plink --bfile chr_1 --bmerge chr_1_par --out chr_1_combined
We now compute the noisy PGI using the observed offspring and parental genotypes:
pgs.py direct_v1_obs --bed chr_@_combined --weights causal_effects.txt --beta_col direct_v1 --pedigree pedigree.txt
To complete the inputs to two-generation PGI analysis, we need an estimate of heritability, as one would obtain from sib-regression, RDR, or MZ-DZ twin comparisons. This estimate is a downard biased estimate of the equilibrium heritability, \(h^2_\text{eq}\), by a factor of \((1-r_\delta)\), where \(r_\delta\) is the correlation between the parents’ direct genetic effect components.
We can obtain this from the VCs.txt output of the simulation, which can be read into R/Python/etc as table. Each row gives, for each generation, the variance of the direct genetic effect component, the phenotypic variance, and the correlation between parents’ direct genetic effect components. The equilibrium heritability is obtained by using the values in the last row: dividing the variance of the direct genetic effect component (first column) by the phenotypic variance (second column). To then obtain the heritability as estimated by sib-regression, RDR, and MZ-DZ twin comparisons, we multiply the equilibrium heritability by \((1-r_\delta)\), where \(r_\delta\) is obtained from the third column of the last row. The equilibrium heritability should be around 0.59, and \(r_\delta\) should be around 0.29, so the heritability as estimated by sib-regression, RDR, MZ-DZ twin comparisons should be around \(h^2_f \approx (1-0.29) \times 0.59=0.42\).
We can now perform two-generation PGI analysis accounting for assortative mating using the following command, with the h2f argument set to the number computed from your VCs.txt file as outlined above (here we use 0.42):
pgs.py direct_v1_obs --pgs direct_v1_obs.pgs.txt --phenofile phenotype.txt --h2f 0.42,0
This script will take the input heritability estimate (0.42) and the standard error of the estimate (here 0 since we used the true value) to estimate the fraction of heritability the PGI would explain in a random mating population, \(k\), which should be around 0.5; the correlation between parents’ direct genetic effect components, \(r_\delta\), which should be around 0.29; the equilibrium heritability, \(h^2_\text{eq}\), which should be around 0.59; the ratio between direct and population effects that would be expected based on assortative mating alone, \(\rho_k\), which should be around 0.86; the indirect genetic effect of true direct genetic effect PGI, \(\alpha_\delta\), which should not be statistically significantly different from zero (with high probability) because there are no parental indirect genetic effects in this simulation; and \(v_{\eta:\delta}\), the contribution to the phenotypic variance from the indirect genetic effect component correlated with direct genetic effect component, which should also not be statistically indistinguishable from zero (with high probability). These estimates are output to direct_v1_obs.am_adj_pars.txt.