Guide
Introduction
snipar (single nucleotide imputation of parents) is a Python package for inferring identity-by-descent (IBD) segments shared between siblings, imputing missing parental genotypes, and for performing family based genome-wide association and polygenic score analyses using observed and/or imputed parental genotypes.
snipar can use any genotyped samples who have at least one genotyped full-sibling or parent.
The imputation method and the family-based GWAS and polygenic score models are described in Young et al. 2022.
Installation
snipar currently supports Python 3.7-3.9 on Linux, Windows, and Mac OSX. We recommend using Anaconda 3 (https://store.continuum.io/cshop/anaconda/).
Installing Using pip
The easiest way to install is using pip:
pip install snipar
Sometimes this may not work because the pip in the system is outdated. You can upgrade your pip using:
pip install –upgrade pip
You may encounter problems with the installation due to Python version incompatability or package conflicts with your existing Python environment. To overcome this, you can try installing in a virtual environment. In a bash shell, this could be done by using the following commands in your directory of choice:
python -m venv path-to-where-you-want-the-virtual-environment-to-be
You can activate and use the environment using
source path-to-where-you-want-the-virtual-environment-to-be/bin/activate
snipar can also be installed within a conda environment using pip.
Installing From Source
To install from source, clone the git repository (https://github.com/AlexTISYoung/snipar), and in the directory containing the snipar source code, at the shell type:
pip install .
Python version incompatibility
snipar does not currently support Python 3.10 or higher due to version incompatibilities of dependencies. To overcome this, create a Python3.9 environment using conda and install using pip in the conda environment:
conda create -n myenv python=3.9
conda activate myenv
pip install snipar
Running tests
To check that the code is working properly and that the C modules have been compiled, you can run the tests using this command:
python -m unittest snipar.tests
Workflow
A typical snipar workflow for performing family-based GWAS (see flowchart below) is:
Inferring identity-by-descent (IBD) segments shared between siblings (ibd.py)
Imputing missing parental genotypes (impute.py)
Estimating direct effects and non-transmitted coefficients (NTCs) of genome-wide SNPs (gwas.py)
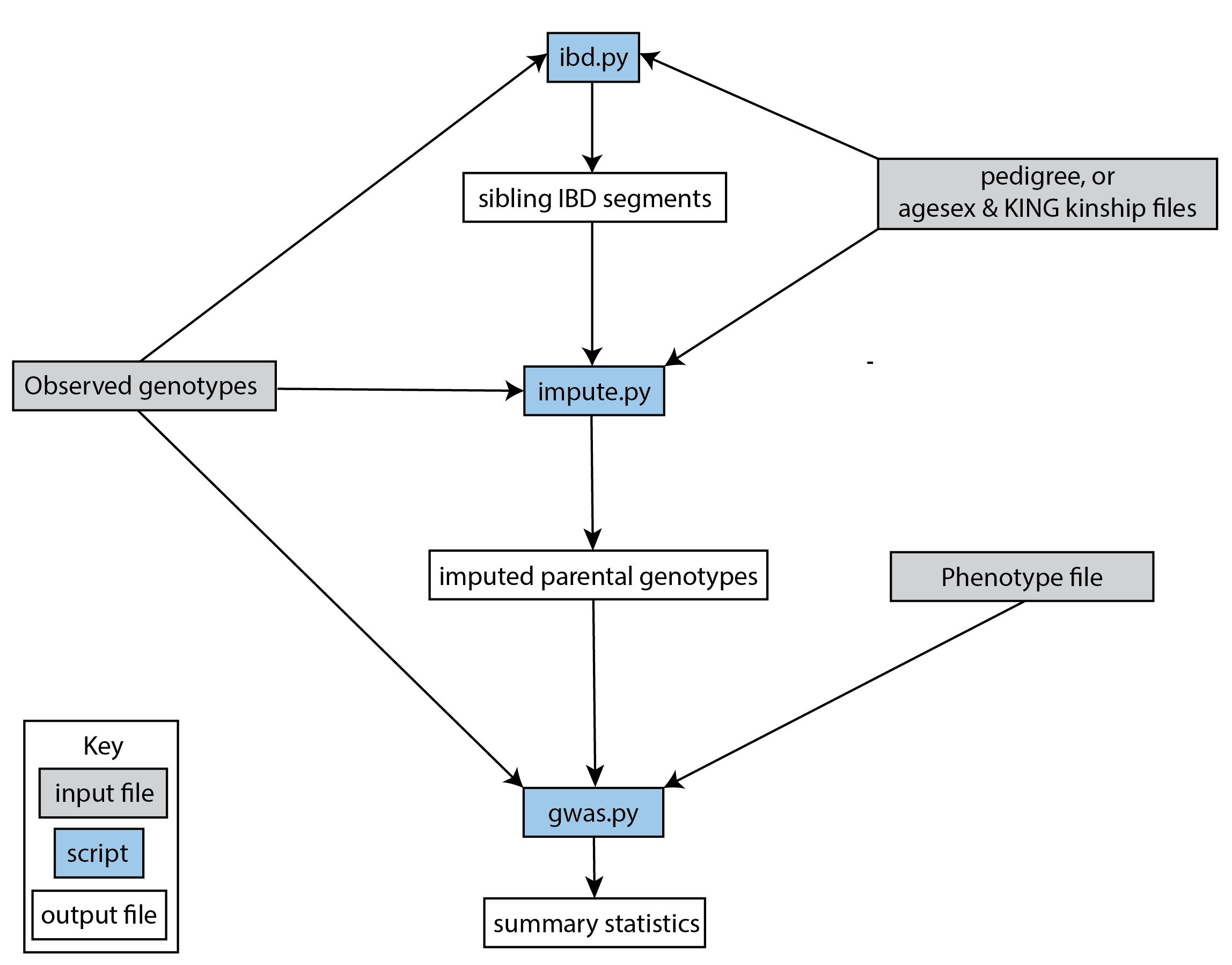
Illustration of a typical workflow for performing family-based GWAS
A snipar workflow requires input files in certain formats. See input files. Output files are documented here.
The tutorial allows you to work through an example workflow before trying real data.
Inputting multiple chromosomes
We recommend splitting up observed genotype files by chromosome since certain scripts in snipar cannot handle observed genotype files with SNPs from multiple chromosomes.
To run scripts for all chromosomes simultaneously (recommended), the @ character can be used as a numerical wildcard. For example, if you had observed genotype files chr_1.bed, chr_2.bed, …, chr_22.bed, then you could specify these as inputs to the command line scripts as “–bed chr_@”. If you only want to analyse a subset of the chromosomes, you can use the “–chr_range” argument; for example, ‘–bed chr_@ –chr_range 1-9’ would specify analysing observed genotype files chr_1.bed, chr_2.bed, …, chr_9.bed.
This will result in output files that are also split by chromosome. The names of the output files can also be specified using the numerical wildcard character, @, e.g. ‘–out /path/to/output/dir/chr_@’.
Inferring identity-by-descent segments
If your sample contains full-sibling pairs (without both parents genotyped), it is necessary to first infer the identity-by-descent (IBD) segments shared between the siblings before imputing the missing parental genotypes. If your sample does not contain any full-sibling pairs, but has genotyped parent-offspring pairs (i.e. one parent’s genotype is missing), imputation can proceed without inferring IBD.
snipar contains a Hidden Markov Model (HMM) algorithm for inferring IBD shared between siblings, which can be accessed through the command line script ibd.py.
The ibd.py script requires the observed genotypes of the siblings and information on the sibling and parent-offspring relations in the genotyped sample.
To infer IBD, one can use a smaller set of genetic variants than one intends to use in downstream analyses (imputation, gwas, etc.). For example, one could use the variants on a genotyping array to infer IBD segments, and these IBD segments could be used to impute missing parental genotypes for a larger set of variants imputed from a reference panel. This can be useful since the accuracy of IBD inference plateaus as the density of variants increases, so inputting millions of variants imputed from a reference panel to ibd.py will result in a long computation time for little gain in accuracy over using variants from a genotyping array.
The information on the relations present in the genotyped sample can be provided through a pedigree file or through the output of KING relationship inference (as output using the –related –degree 1 options: see https://www.kingrelatedness.com/manual.shtml#RELATED) along with a file giving the age and sex information on the genotyped sample. (The age and sex information along with the parent-offspring and sibling relations inferred by KING are used to construct a pedigree if a pedigree is not provided.)
The algorithm requires a genetic map to compute the probabilities of transitioning between different IBD states. If the genetic map positions (in cM) are provided in the .bim file (if using .bed formatted genotypes), the script will use these. Alternatively, the –map argument allows the user to specify a genetic map in the same format as used by SHAPEIT (https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html#formats). If no genetic map is provided, then the deCODE sex-averaged map on GRCh38 coordinates (Halldorsson, Bjarni V., et al. “Characterizing mutagenic effects of recombination through a sequence-level genetic map.” Science 363.6425 (2019).), which is distributed as part of snipar, will be used.
The HMM employs a genotyping error model that requires a genotyping error probability parameter. By default, the algorithm will estimate the per-SNP genotyping error probability from Mendelian errors observed in parent-offspring pairs. However, if your data does not contain any genotyped parent-offspring pairs, then you will need to supply a genotyping error probability. If you have no external information on the genotyping error rate in your data, using a value of 1e-4 has worked well when applied to typical genotyping array data.
The HMM will output the IBD segments to a gzipped text file with suffix ibd.segments.gz. As part of the algorithm, LD scores are calculated for each SNP. These can also be output in LDSC format using the –ld_out option.
Imputing missing parental genotypes
impute.py is responsible for imputing the missing parental genotypes. This is done for individuals with at least one sibling and/or parent genotyped but without both parents genotyped.
You should provide the script with identity-by-descent (IBD) segments shared between the siblings if there are genotyped sibling pairs in the sample. Although we strongly recommend using IBD segments inferred by ibd.py, we also support IBD segments in the format that KING outputs (see https://www.kingrelatedness.com/manual.shtml#IBDSEG). If IBD segments in KING format are used, it is necessary to add the –ibd_is_king flag.
The script needs information about family structure of the sample. You can either supply it with a pedigree file or let it build the pedigree from kinship and agesex files.
If you are imputing for a chromosome with a large number of SNPs, you may encounter memory issues. If this is the case, you can use the –chunks argument to perform the imputation in chunks. When the script is run with ‘-chunks x’, it will split the imputation into ‘x’ batches. Alternatively, you can do the imputation for only on a subset of SNPS by using -start and -end options.
For each chromosome, imputed parental genotypes and other information about the imputation will be written to a file in HDF5 format. The contents of the HDF5 output, which a typical user does not need to interact with directly, are documented here.
The expected proportion of variants that have been imputed from a sibling pair in IBD0 (i.e. the parental alleles are fully observed) can be computed from the pedigree. At the end of the imputation, the script will output the expected IBD0 proportion and the observed IBD0 proportion. If there have been issues with the imputation (such as failure to match IBD segments to observed genotypes), this will often should up as a large discrepancy between expected and observed IBD0 proportions.
Family-based genome-wide association analysis
Family-based GWAS is performed by the gwas.py script. This script estimates direct effects, non-transmitted coefficients, and population effects of input genetic variants on the phenotype specified in the phenotype file. (If multiple phenotypes are present in the phenotype file, the phenotype to analyse can be specified using the ‘–phen_index’ argument, where ‘–phen_index 1’ corresponds to the first phenotype.)
The script will use both observed and imputed parental genotypes to estimate these effects. Note that if no imputed parental genotypes are input, effects will be estimated using individuals with both parents genotyped only, provided that a pedigree file is also input. (A pedigree input is not needed when inputting imputed parental genotypes.)
By default, for each variant, the script performs a regression of an individual’s phenotype onto their genotype, their (imputed/observed) father’s genotype, and their (imputed/observed) mother’s genotype. This estimates the direct effect of the variant, and the paternal and maternal non-transmitted coefficients (NTCs). See Young et al. 2022 for more details.
If no parental genotypes are observed, then the imputed maternal & paternal genotypes become perfectly correlated. In this case, to overcome collinearity, gwas.py will perform a regression of an individual’s phenotype onto their genotype, and the imputed sum of their parents’ genotypes. This will estimate the direct effect of the SNP, and the average NTC.
If one wishes to model indirect genetic effects from siblings, one can use the ‘–fit_sib’ option to add the genotype(s) of the individual’s sibling(s) to the regression.
The gwas.py script first estimates a variance component model that models the phenotypic correlation between siblings, then does a transformation that allows the SNP effects to be estimated by simple linear regression while accounting for correlations between siblings.
The script outputs summary statistics in both gzipped text format and HDF5 format.
Estimating correlations between effects
As part of Young et al. 2022, we estimated the genome-wide correlations between direct and population effects and between direct effects and average non-transmitted coefficients (NTCs). The correlation between direct effects and population effects is a measure of how different direct effects and effects estimated by standard GWAS (population effects) are.
We provide a script, correlate.py, that estimates these correlations. It takes as input the summary statistics files output by gwas.py and LD-scores for the SNPs (as output by ibd.py or by LDSC). It applies a method-of-moments based estimator that accouts for the known sampling variance-covariance of the effect estimates, and for the correlations between effect estimates of nearby SNPs due to LD.
Note that this is different to genetic correlation as estimated by LDSC. LDSC attempts to use LD-scores to estimate heritability and to separate out this from bias due to population stratification. The correlate.py estimator only uses LD-scores to account for correlations between nearby SNPs, not to separate out population stratification. This is because we are (potentially) interested in the contribution of population stratification to population effects, and whether population stratification makes population effects different from direct effects. The approach used by LDSC would remove some of the contribution of population stratification to differences between direct and population effects.
Family-based polygenic score analyses
As in previous work (e.g. Kong et al. 2018: https://www.science.org/doi/abs/10.1126/science.aan6877), parental polygenic scores can be used as ‘controls’ to separate out the component of the association between phenotype and polygenic score (PGS) that is due to direct genetic effects. In Young et al. 2022, we showed how this can be done using parental PGSs computed from imputed parental genotypes. snipar provides a script, pgs.py, that can be used for computing and analysing PGSs using observed/imputed parental genotypes.
The pgs.py script takes similar inputs to the gwas.py script. The main addition is that in order to compute a PGS, a weights file must be provided.
By default, if no phenotype file is provided, the pgs.py script will compute the PGS values of all the genotyped individuals for whom observed or imputed parental genotypes are available. The script will output a PGS file, including the imputed/observed PGS values for each individual’s parents, facilitating family-based polygenic score analyses.
If the ‘–fit_sib’ argument is provided, the PGS file will include a column corresponding to the average PGS value of the individual’s sibling(s).
To estimate the direct and population effects as well as the non-transmitted coefficients (NTCs) of the PGS on a phenotype, input a phenotype file to pgs.py. One can first compute the PGS and write it to file, and then use this as input to pgs.py along with a phenotype file.
The direct effect and NTCs of the PGS are estimated as fixed effects in a linear mixed model that includes a random effect that models (residual) phenotypic correlations between siblings. The population effect is estimated from a separate linear mixed regression model that includes only the proband PGS as a fixed effect. The estimates and their standard errors are output to file along with a separate file giving the sampling variance-covariance matrix of the direct effect and NTCs.